Section: New Results
Statistical Characterization, Modelling and Classification of Morphological Changes in imp Mutant Drosophila Gamma Neurons
Participants : Agustina Razetti, Caroline Medioni, Florence Besse, Xavier Descombes.
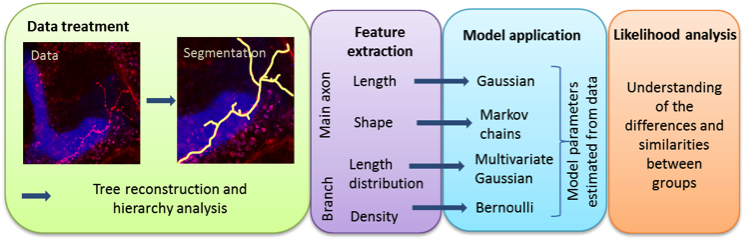
In Drosophila brain, gamma neurons in the mushroom body are involved in higher functions such as olfactory learning and memory. During metamorphosis, they undergo remodelling after which they adopt their adult shape. Some mutations alter remodelling and therefore neuronal final morphology, causing behavioural dysfunctions. The RNA binding protein Imp, for example, was shown to control this remodelling process at least partly by regulating profilin expression. This work aims at precisely characterizing the morphological changes observed upon imp knockdown in order to further understand the role of this protein. We developed a methodological framework that consists in the selection of relevant morphological features (axon length and shape and branch length distribution and density), their modelling and parameter estimation. We thus perform a statistical comparison and a likelihood analysis to quantify similarities and differences between wild type and mutated neurons. The data was a set of 3D images showing a single neuron taken with a confocal microscope and provided by F. Besse group, IBV. The workflow from raw data to the likelihood analysis is summarized on figure6 . We show that imp mutant neurons can be classified into two phenotypic groups (called Imp L and Imp Sh) that differ in several morphological aspects. We also demonstrate that, although Imp L and wild-type neurons show similarities, branch length distribution is discriminant between these populations. Finally, we study biological samples in which Profilin was reintroduced in imp mutant neurons, and show that defects in main axon and branch lengths are partially suppressed.